
These reagents often lower the primer T m, so the annealing temperature should be adjusted accordingly. To overcome strong GC interactions, the most common approach relies on PCR additives or co-solvents such as DMSO to help DNA denature ( Figure 6A). To amplify GC-rich targets, the double-stranded template must be separated for the primers to bind and DNA polymerase to read through the sequence. Thus, GC-rich sequences can cause DNA polymerases to “stutter” along templates and interrupt DNA synthesis. GC-rich sequences can also be involved in secondary structures. In this manner, desired PCR products are selectively increased with little or no amplification of nonspecific targets over the course of PCR ( Figure 2).ĭNA templates containing high GC content (>65%) can be difficult to amplify because of the stronger hydrogen bonds between G and C bases. Once the annealing temperature reaches, or “touches down”, at the optimal temperature (usually 3–5☌ lower than the lowest primer T m), it is maintained throughout the remaining cycles for primer annealing. To overcome this challenge, the annealing temperature is often decreased 1☌ at every cycle of the initial few cycles to produce a sufficient yield of the desired amplicon. While preventing primer-dimers and nonspecific primer binding, the higher annealing temperatures may result in lower PCR yield due to increased dissociation of primers from their intended target. As such, higher annealing temperatures reduce nonspecific PCR products and promote specific amplification at the start of PCR (learn more about PCR annealing step). Higher temperatures help destabilize the formation of primer-dimers and nonspecific primer-template complexes, thus minimizing undesirable amplification. In touchdown PCR, the annealing temperature of the first few cycles is set to be a few degrees higher than the highest melting temperature (T m) of the primers. NOT FOR USE IN DIAGNOSTIC PROCEDURES (EXCEPT AS SPECIFICALLY NOTED).Another approach to promoting specificity is to modify the PCR cycling parameters. Our mission is to develop high-quality innovative tools and services to accelerate discovery.įOR RESEARCH USE ONLY. As a member of the Takara Bio Group, Takara Bio USA is part of a company that holds a leadership position in the global market and is committed to improving the human condition through biotechnology. provides kits, reagents, instruments, and services that help researchers explore questions about gene discovery, regulation, and function. This domain swap PCR was designed to insert the IGHV gene into the vector by PCR using PrimeSTAR GXL DNA Polymerase and two overlapping primers. IGHV4-59 was then mixed with the expression vector DNA for a domain swap quick-change PCR. To clone IGHV4-59 into a larger expression vector (approximately 9 kb), IGHV4-59 was re-amplified and purified for use as a megaprimer. This cDNA was used as template for amplification of the IGHV4-59 gene by PCR with PrimeSTAR GXL DNA Polymerase. In brief, total RNA from human PBMC cells was reverse transcribed to synthesize cDNA. This approach required a high-fidelity PCR polymerase to re-amplify a larger plasmid vector with insert cDNA. The objective of this experiment was to amplify a low-expression antibody heavy-chain variable region gene (IGHV4-59) from human B cells and clone it into an expression vector using a domain-swap megaprimer PCR technique.
#TOUCHDOWN PCR TO AMPLIFY 3KB PLASMID REGION SERIES#
Therefore, megaprimer PCR is an alternative to conventional restriction enzyme digestion and ligation for the construction of individual clones or a series of chimeric clones.
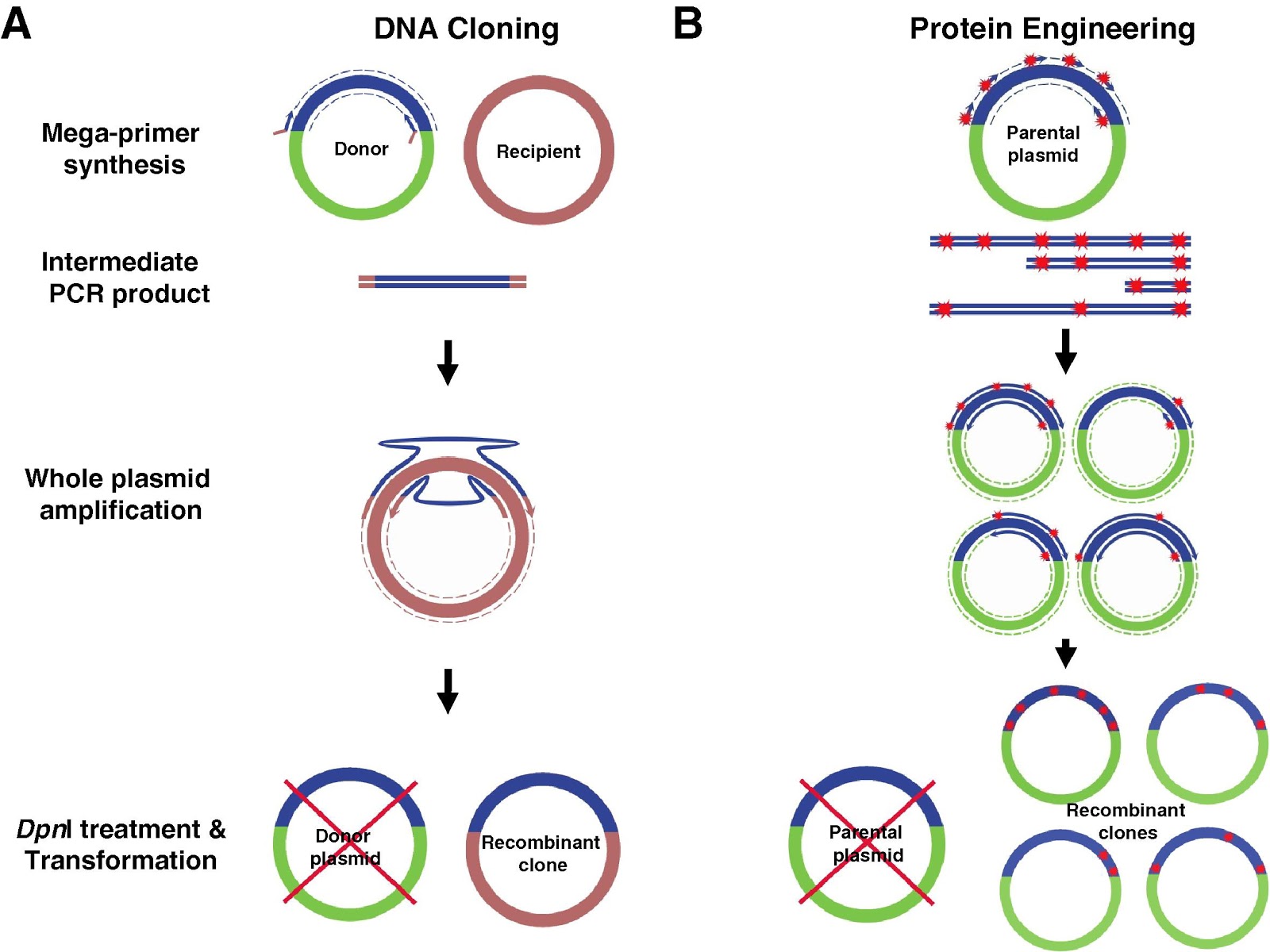
This second PCR may be designed to allow domain swapping and synthesis of entire plasmids. Megaprimer-mediated domain swapping involves construction of clones using PCR to synthesize DNA fragments, then using those fragments as megaprimers for a second PCR.


 0 kommentar(er)
0 kommentar(er)
